PopRep 1.0 User's Manual
Eildert Groeneveld
Department of Animal Breeding and Genetics
Institute of Farm Animal Genetics (FLI)
Mariensee, Germany
Preface
Welcome to POPREP! Pedigrees are abundent all around us: canary bird and cat breeders are convinced of their importance, humans sometimes pride themselves of a distinguished pedigree, dog chow uses its name. Pedigrees are funny animals (in dioecious species): their individual elements for one generation are simple: the name of a being, its father and mother, its sex and perhaps a birthdate to place it into the time dimension. But then come the more exciting characteristics: pedigrees are recursive: a father is a being himself and has a father and mother which again has the same property, leading us into the metaphysical realm: where does it all end (as such recursions do not end) and who was the mother of that first one? The number of ancestors doubles from one generation to the next, producing a nice tree that grows like no tree does. Yet we know, that all started with only the two: Adam and Eve (which leads us to one important aspect in POPREP, namely inbreeding).
POPREP will not go back as far as the big bang or Adam and Eve but rather dwell upon the intermediate between now and then. The reader will be surprised how much can be deduced from an innocently looking pedigree. And to make this possible with appropriate ease, starting such an evaluation does not require a download (and installation) of a software system but instead an upload of data through the internet.
Here the recursion of life meets the unidirectional development in computing: sending data into the cloud and receiving the results back from the sky, which, hopefully, allow tackling down to earth problems.
This document is targeted at end users of POPREP to supply them with the necessary information to setup the input data files and to interpret the outputs. Furthermore, it outlines the options for postprocessing of results and those for customizing printed outputs.
In a separate section also installation issues of the software and the demands on the computing hardware are outlined. This part adresses the system administrator.
1. Introduction
Conservation of breeds under the threat of extinction has long been an issue of importance. In the wake of the Rio Convention [6], the focus of conservation has expanded to within breed diversity of small but also large populations which are - as such - not under the threat of extinction. The reasons for concern lie in the high use of few genetically superior males as made possible by artificial insemination resulting in increased rates of inbreeding. The Holstein breed, for example, in terms of absolute numbers is one of the most abundant breeds, but it may be loosing genetic diversity at an undesirable rate as indicated by its rather low effective population size [5]. While management of biodiversity within small populations is of importance, it is even more relevant in large mainstream populations. As they are responsible for much of the food production their deterioration would have a wide spread negative effect. Therefore, the National Action Program on Animal Genetic Resources in Germany [2] stipulates continuous monitoring of active breeding populations. Accordingly, management of populations that are not under direct threat of extinction has also become an issue.
Often good information on generation intervals, rates of inbreeding and effective population sizes as required for population management is not available. In cooperative breeding programs selection decisions are usually taken by individual breeders. The degree of utilization of sires results from the sum of individual breeders' decisions. Then, the cumulative effect of individual selection decisions can only be assessed ex post. However, if the trends were known early on, counteractive measures could still be taken.
It is the objective of this contribution to develop a generic report and implement it in software based on a minimum set of individual animal information. The intended use is threefold: firstly, it can serve as a documentation of parameters relevant to biodiversity issues to be included in the yearly reporting on population as proposed in [3]. Secondly, its outputs can serve as an early warning system in the management of big and small populations allowing to counteract negative trends as soon as they become apparent. Finally, intermediate output will be made available for further reseach.
POPREP is a software package which computes on the basis of pedigrees a number of statistics that can be used in the management of breeding populations, helping to ascertain the current status of a given population in a number of terms that are related to biodiversity issues.
2. Installation of PopRep
A number of options exist for the use of POPREP.
- Being an APIIS application POPREP come as an integral part of any such APIIS installation and can be run against the actual database.
- use of RapidAPIIS to create a customized local datadase
- the third option is the POPREP web site which is maintained by the Institute of Farm Animal Genetics (popreport.fli.de).
In the following we shall assume the use of the POPREP website (popreport.fli.de).
3. Popreport.fli.de
The POPREP software is installed on the compute servers at the Institute of Farm Animal Genetics (FLI) as resource offered to the scientific community in the management and assessment of biodiversity.
Instead of transfering the software to the users and having them install everything on their own computer, here the data is transfered to the POPREP website, where the computations are executed. Upon completion the results are mailed back to the user and the data is erased from the server.
3.1 Interacting with Popreport
Accordingly, the user interface to PopRep is simple and requires only 6 inputs (see Figure 1): the email address to which the results will get sent, the name of the breed or population that the user wants to show up in the reports, the code used for male and female (e.g. m,f or 1,2) in the data and the representation of the birthdate, a possible date offset, and the upload of the pedigree file to the web site. Furthermore, the user may check a box if apart from the reports provided in pdf format also intermediate results are to be provided. In that case additional to the reports an archive of intermediate results files will be added to the mail.
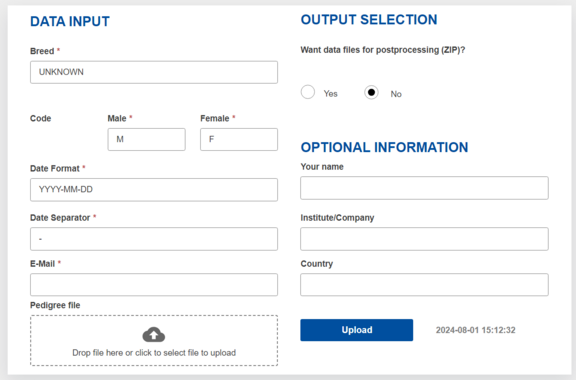
After upload the user is informed if the job has been submitted right away for processing or if it is place in the waiting queue because of other jobs being currently processed.
In due course (which may be minutes or hours, depending on the load on the system and the size of the problem), the user will find the results in the mail box.
3.2 Minimal data set required for Popreport
Data to be analyzed by POPREP have to be supplied in one ASCII file adhering to the format specifications below. The columns of this file are:
- animal ID (not null)
- sire ID (may be null)
- dam ID (may be null)
- Date of birth of animal (may be null)
- sex of animal (not null)
These five columns are delimited by the pipe sign: | and accordingly do have to be in a fixed column format. However, columns do not have to be alligned. An example is given in Table 1. In setting up this file the following issues have to be considered:
IDs:
the animal sire and dam ID need to be considered as one pool of identifications. This means, that all have to have the same format. For each sire and dam an animal record needs to exist with the identical identification string. If a sire or dam is unknown its corresponding ID is the string 'unknown'. Alternatively, for a base animal listed as sire or dam an own animal record of such a base animal can be omitted (and will be automatically generated by PopRep). Note, that in the latter case, the birthdate of the base animal can not be specified while in the case of an own animal record of a base animal this option does indeed exist.
Alpha character as part of the ID are considered case sensitive. This means that 'DE100212' and 'de100212' is not the same! Along the same lines '0123' is not the same as '123'. Users should always view the ID as character strings even if they are always numerical. Blanks cannot be part of the ID strings!
The animal ID must not be left unknown or blank. Sires and dams, on the other hand may contain either |unknown| or || . Instead of 'unknown' the user can also use 'unknown_sire' and 'unknown_dam' in the sire and dam column, respectively, as shown in Table 1.
Birthdate:
much of the computations is done on the basis of the birthdate, such as number of sires and dams used or generation intervals. When birthdates are not available, as may be the case for base animals, then these records will be skipped in these computations, whereas in global computations like for the inbreeding levels of individual animals no birthdate is required. Unknown birth dates are represented by two consecutive pipes: ||.
Sex:
dams and sires are evaluated separately, this is why the sex of the animals need to be known. Here, only intact males and females should be included. For this column, no unknown (empty) fields are allowed!
For all strings: the user should take care to avoid special characters which could produce problems somewhere in the chain of computing. Thus, if unexpected results occur, the user may want to consider this.
Two approaches can be chosen regarding the data selection for the pedigree file. In an animal recording scheme that collects birth records along with the identification for the new born animals, the animal register will contain all animals born in the population plus those that were used as sires and dams. Other datasets may just comprise selected sires and dams. Apart from the volume differences of the two types, the outputs generated will be different.
3.3 Input validation
Prior to data validation the email adress is check for validity. After uploading of the pedigree file, its format and content is checked for validity. This starts with the requirement of having pipe delimited columns as shown in Table 1.

3.4 Computational issues
Behind the scene of one POPREP run quite a number of independent tasks are executed. The validation procedure has already been described above. Thereafter, a Postgresql database is created and the pedigree data loaded. The figures for the population structure report are generated by SQL commands on the database. Furthermore, for the inbreeding report an number of other software components are involved. Perl is used to compute the individual animal inbreeding coefficients, as is the pedigree completeness. However, the latter is parallelized in that 6 processes are started in parallel, using all the 6 processors of the WEB server at the same time on one report. The other big block of computations is the average genetic relationship among the animals of each birth year. This is done through a FORTRAN program using Didier Boichards pedig software. This is also done in parallel using all available processors on the server. Finally, LATEX and gnuplot are used to typeset the final PDF reports.
Depending on the load and size of the problem, the reports can be produced within minutes up to hours. Just be patient, at some point you will get an email telling you what has happened and hopefully a bunch of valuable population reports with figures you have never seen before.
3.5 Server house keeping
As has been stated above any user with a valid email address can upload data to the server. The pedigrees can tell a lot about the breeding population and, thus, the user will not want this to be public. To understand the security related issue, the following gives the sequence of events executed on the PopRep server upon pressing the upload button on a user's browser:
- WEB forms check:
- email address checked for validity
- date format and date separator checked for consistency and validity
- field 'Breed' checked for illegal characters (blanks, LATEX special characters)
- log entry of user created on the basis of the web form content
- data uploaded and stored in a temporary file on the server
- new APIIS project created; new database user created
- database created
- prechecking of data executed
- data loaded into the newly created DB
- computations executed against the database
- results checked for completeness and mailed back to the user
- database destroyed
- upload files and results deleted
- user log completed and saved
As can be seen, the after processing uploaded data plus the generated reports and intermediate results are deleted. Therefore, if you somehow deleted the results on your computer or if it got lost during the emailing, there is no point in writing us to provide the mail again: we do not have it. Then you will have to rerun the dataset.
3.6 Receiving the results
The results from a POPREP run are returned through email to the initiating user. The minimum to be returned is the type set report on the uploaded pedigree of one breed or breeding population. Typically, this is a 40 page document in pdf format of around 1MB, that the user can print on the local printer. If during upload of data the user chooses the compressed archive of numerical intermediate data, the mail may get substantially bigger, in particular for a large pedigree.
4. Postprocessing
The default output from POPREP are the two typeset PDF documents: population structure report and population inbreeding report. These are selfcontained documents ready to be printed. Each chapter has an in depth explanation of computational methods used and how the tables/graphs can be interpreted. If the post processing box is ticked in the web user interface, then the numerical values that got used to generate the reports will also be mailed to the user. For each table a csv (comma separated value) file is provided. In this way, the user can load this data into her favourite spreadsheet software and analyse the data further.
The content of the pedigree file is validated next. There are two classes of error: the first leads to an abort of the processing while the second set of errors leads to the user notification but continues with the data processing.
job terminating errors:
- Illegal input file format:
as listed above 5 data columns are assumed in the input file, the columns delimited by the pipe (|) sign. Furthermore, the date format needs to comply to the specifications. If the input does not conform, a corresponding message is written and further processing stopped. - Loops
in pedigrees always lead to an abort of data processing, accordingly, no population report will be produced. A pedigree loop occurs if at some point in the pedigree an animal ID is listed as its own ancestor. In the computation of inbreeding this would lead to an infinite loop, thus, this condition is part of the initial input validation. Members of the loop are printed in the run log which in turn is mailed to the submitting user. It is then the obligation of the user to fix the input data and resubmit the job.
Notice, that the process get stopped after the first loop has been found. Thus, with multiple loops in the pedigree file, the user may need to upload and start the process repeatedly.
non job terminating errors:
- sire and dam sex consistency:
in column 5 the sex of the animal is specified. Implicitly, column 2 and 3 refer to males and females, respectively. This check verifies, that sires only show up as male animals in the pedigree and dams only as females. Again, the process gets aborted and an error message sent to the user. - sire and dam consistency:
clearly, the sire ID must not equal the dam ID; here, no action is taken, i.e. the wrong record remains in the database. Again, the user should fix the problem and rerun. - ancestor birthdate
must not be larger than that of the offspring. This is the default when the user in the WEB GUI does not specify the minimum age of a parent at birth of the first offspring; then the default is set to 0, else this offset is added to the birthdate of the parent prior to making the comparison to its offspring. There is no clear resolution how to handle violations: either the offspring birthdate or that of the parent may be wrong. However, we have decided to take action so that the report can be generated: the birthdates of the conflicting parties are all set to unknown. The effects of this action is, that the animals for with the birthdate was set to unknown will not be included in the cohort definitions which tend to operate on the year of birth, while they will be included in the computation of the inbreeding coefficients but not in the additive genetic relationships. It is advisable, that the user cleans the data and runs the job again.
Should there be any errors in the input file identified by POPREP the user should fix those and rerun. There is not much worse, than publishing results based on input data errors. Furthermore, the user must be aware, that not all errors in the data can be detected. Clearly, wrong sire identifications can not be found by any software. Even with extensive testing through PopRep it is the user's responsibilty to provide correct data.
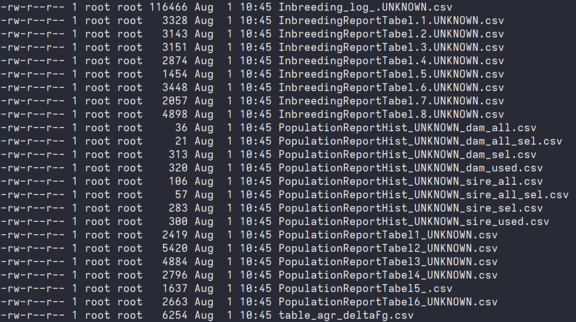
As can be seen from the file names, most of them correspond to the table and figures in the printed reports and can easily be identified. As can been seens, all these files are small with just a few hundred bytes. The file that sticks out is the one starting with ``Inbreeding_log''.
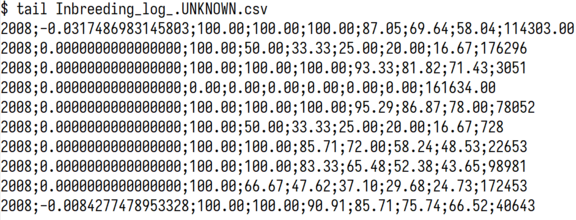
Table 3 shows one line for each animal in the pedigree file with the birth year in column 1. Then follows the \(ln(1-\Delta F_{i})\) for that animal and the degree of pedigree completeness for 1 through 6 generations backward, followed by the animal identification. As an example, the last entry is for an animal born in the year 2008 with an \(ln(1-\triangle F_{i})=-.0084277\) , with complete pedigree going 1 and 2 generations back, and 91%, 86% and 76% for pedigrees 3, 4, and 5 back.
5. Computational Methods
The computational issues are described in detail in the paper ``POPREP: A Generic Report for Population Management'', E. Groeneveld, B. v.d. Westhuizen, A. Maiwashe, F. Voordewind and JBS. Ferraz, Genetics and Molecular Research (GMR), 8(3), to be published September 15, 2009.
6. Implications of data selection
As can be seen from the WEB interface, very little can be done by the user other than uploading the dataset to POPREP. The choice of the dataset and the selection of records is ofcourse under the control the user. Anything POPREP computes is, ofcourse, only a function of the input data. Not only do data errors influence the results (described above) but also the selection of the pedigree data itself (see 3.2).
7. Acknowledgement
Only through the dedication of a number of persons has it beed possible to develop the POPREP package and the WEB site now open for general use. These are:
- Frits Voordewind: PERL/SQL/GNU-Plot/Report, ARC, Irene, South Africa
- Bobbie van der Westhuizen: PERL/SQL/GNU-Plot/Report, ARC, Irene, South Africa
- Norman Maiwashe: Report/Descriptions, ARC, Irene, South Africa
- Ralf Fischer: Computation of Inbreeding, LfULG , Köllitsch, Germany
- Didier Boichard: PEDIG software, INRA, France[1]
- Lina Yordanova: SQL, University of Stara Zagora, Bulgaria
- Helmut Lichtenberg: Integration and WEB service, FLI, Germany
- Zhivko Duchev: pedigree recursion for PCI, FLI, Germany
- Eildert Groeneveld: Project Leader, FLI, Germany
Furthermore, financial support from the Federal Ministry of Food, Agriculture and Consumer Protection, Germany (BMELV), the Agricultural Research Council of South Africa (ARC) and the German Academic Exchange Service (DAAD) is gratefully acknowledged.
Whenever POPREP used, the following paper should be cited [4]: Eildert Groeneveld, Bobbie v.d. Westhuizen, Azwihangwisi Maiwashe, Frits Voordewind, and José Bento S. Ferraz. Population Management. 2009 submitted. POPREP: A Generic Report for Genetics and Molecular Research (GMR), 8(3)
8. Appendix
The appendix contains a complete sample population report
Bibliography
- Didier Boichard.
Pedig: a fortran package for pedigree analysis suited for large populations.
In Proceedings of the 7th World Congress on Genetics applied to Livestock Production (WCGALP), Montpellier, France, 19.-23. August 2002.
CD-ROM communication No. 28-13. - DGfZ-Ausschuss zur Erhaltung genetischer Vielfalt bei landwirtschaftlichen Nutztieren.
Nationalen Fachprogramm zur Erhaltung und Nutzung tiergenetischer Ressourcen, 2002. - Eildert Groeneveld.
Strategie und Logistik zur verantwortungsvollen Verwaltung der genetischen Diversität in der Nutztierzüchtung.
Züchtungskunde, 75(5):317-323, 2003.
ISSN 0044-5401. - Eildert Groeneveld, Bobbie v.d. Westhuizen, Azwihangwisi Maiwashe, Frits Voordewind, and José Bento S. Ferraz.
POPREP: A Generic Report for Population Management.
Genetics and Molecular Research (GMR), 8(3):??, 2009 accepted. - L. Maignel, Didier Boichard, and Etienne Verrier.
Genetic variability of French dairy breeds estimated from pedigree information.
In Interbull Bulletin, volume 14, pages 49-54. 1996. - United Nations.
United Nations Environment Programme Convention on Biological Diversity.
www.cbd.int, 1992.


